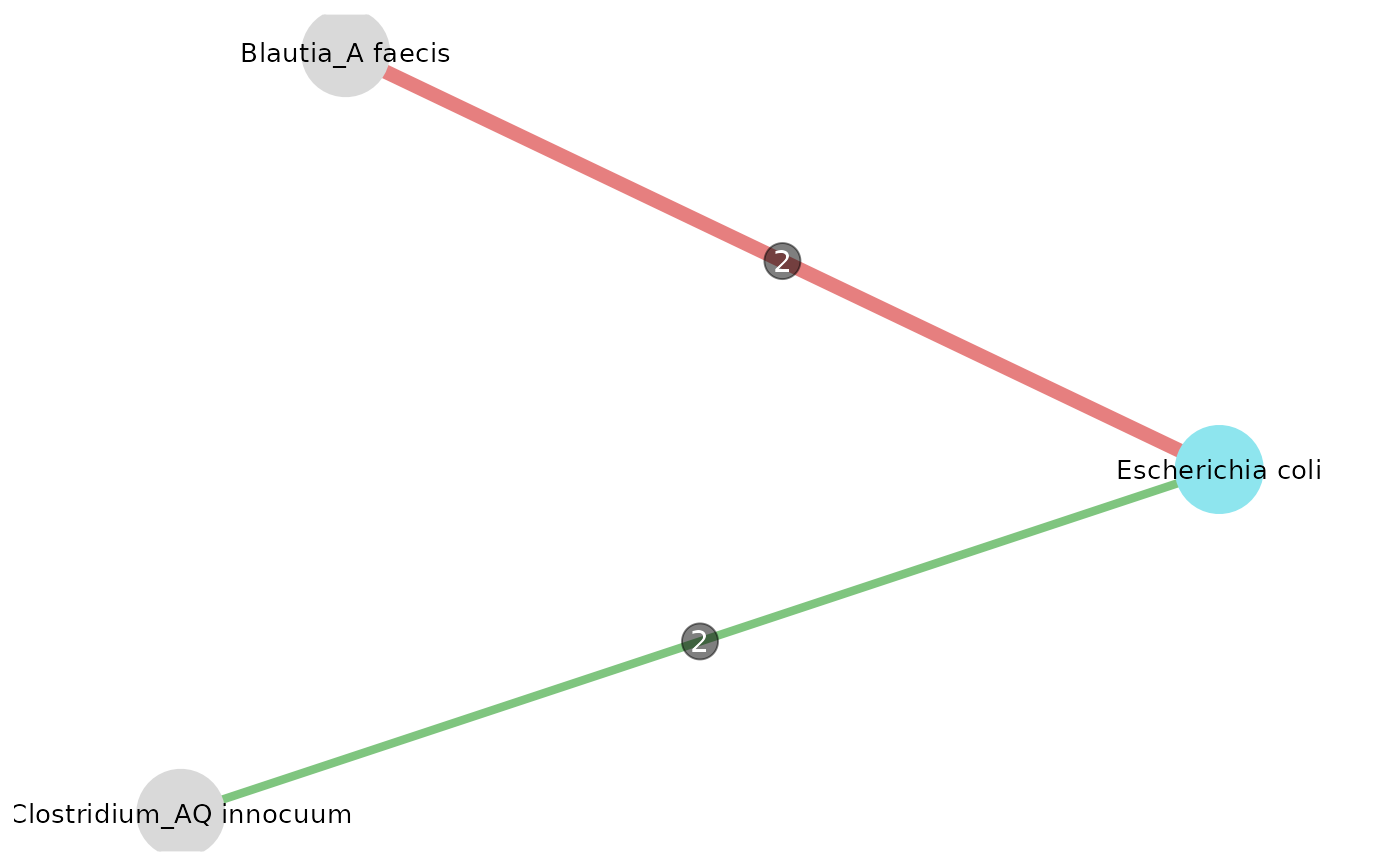
Display the intersection network from 2 or more datasets
intersections_network.RdDisplay the intersection network from 2 or more datasets
Usage
intersections_network(
res_list,
threshold,
annotation_table,
col_module_id,
annotation_level,
object_of_interest,
annotation_option = FALSE,
node_size = 12,
label_size = 4,
edge_label_size = 2,
object_color = "cadetblue2",
seed = NULL
)Arguments
- res_list
List of dataframes. The results from apply_NeighborFinder() on several datasets
- threshold
Numeric. Integer corresponding to the minimum number of datasets in which you want neighbors to have been found
- annotation_table
Dataframe. The dataframe gathering the taxonomic or functional module correspondence information
- col_module_id
String. The name of the column with the module names in annotation_table
- annotation_level
String. The name of the column with the level to be studied. Examples: species, genus, level_1
- object_of_interest
String. The name of the bacteria or species of interest or a key word in the functional module definition
- annotation_option
Boolean. Default value is False. If True: labels on nodes become module names instead of module IDs
- node_size
Numeric. The parameter to adjust size of nodes
- label_size
Numeric. The parameter to adjust size of labels
- edge_label_size
Numeric. The parameter to adjust the size of edge labels
- object_color
String. The name of the color to differentiate the nodes corresponding to 'object_of_interest' from the other module IDs
- seed
Numeric. The seed number, ensuring reproducibility
Examples
data(taxo)
data(data)
data(metadata)
res_CRC_JPN <- apply_NeighborFinder(data$CRC_JPN, object_of_interest = "Escherichia coli", col_module_id = "msp_id", annotation_level = "species")
res_CRC_CHN <- apply_NeighborFinder(data$CRC_CHN, object_of_interest = "Escherichia coli", col_module_id = "msp_id", annotation_level = "species", covar = ~study_accession, meta_df = metadata$CRC_CHN, sample_col = "secondary_sample_accession")
res_CRC_EUR <- apply_NeighborFinder(data$CRC_EUR, object_of_interest = "Escherichia coli", col_module_id = "msp_id", annotation_level = "species", covar = ~study_accession, meta_df = metadata$CRC_EUR, sample_col = "secondary_sample_accession")
intersections_network(res_list = list(res_CRC_JPN, res_CRC_CHN, res_CRC_EUR), taxo, threshold = 2, "Escherichia coli", col_module_id = "msp_id", annotation_level = "species", label_size = 7, edge_label_size = 4, node_size = 15, annotation_option = TRUE, seed = 3)